About the CoMSES Model Library more info
Our mission is to help computational modelers develop, document, and share their computational models in accordance with community standards and good open science and software engineering practices. Model authors can publish their model source code in the Computational Model Library with narrative documentation as well as metadata that supports open science and emerging norms that facilitate software citation, computational reproducibility / frictionless reuse, and interoperability. Model authors can also request private peer review of their computational models. Models that pass peer review receive a DOI once published.
All users of models published in the library must cite model authors when they use and benefit from their code.
Please check out our model publishing tutorial and feel free to contact us if you have any questions or concerns about publishing your model(s) in the Computational Model Library.
We also maintain a curated database of over 7500 publications of agent-based and individual based models with detailed metadata on availability of code and bibliometric information on the landscape of ABM/IBM publications that we welcome you to explore.
Displaying 10 of 10 results predator-prey clear search
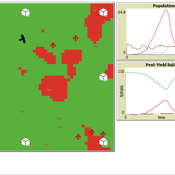
Peer reviewed Avian pest control: Yield outcome due to insectivorous birds, falconry, and integration of nest boxes.
David Jung | Published Monday, November 13, 2023 | Last modified Sunday, November 19, 2023The model aims to simulate predator-prey relationships in an agricultural setting. The focus lies on avian communities and their effect on different pest organisms (here: pest birds, rodents, and arthropod pests). Since most case studies focused on the impact on arthropod pests (AP) alone, this model attempts to include effects on yield outcome. By incorporating three treatments with different factor levels (insectivorous bird species, falconry, nest box density) an experimental setup is given that allows for further statistical analysis to identify an optimal combination of the treatments.
In light of a global decline of birds, insects, and many other groups of organisms, alternative practices of pest management are heavily needed to reduce the input of pesticides. Avian pest control therefore poses an opportunity to bridge the disconnect between humans and nature by realizing ecosystem services and emphasizing sustainable social ecological systems.
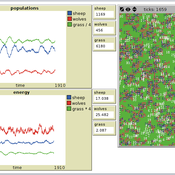
Using a simple ABM to help undergraduates understand impacts of an invasive species: fish, lionfish, and zooplankton
Samantha Farquhar | Published Friday, February 24, 2023This model is an implementation of a predator-prey simulation using NetLogo programming language. It simulates the interaction between fish, lionfish, and zooplankton. Fish and lionfish are both represented as turtles, and they have their own energy level. In this simulation, lionfish eat fish, and fish eat zooplankton. Zooplankton are represented as green patches on the NetLogo world. Lionfish and fish can reproduce and gain energy by eating other turtles or zooplankton.
This model was created to help undergraduate students understand how simulation models might be helpful in addressing complex environmental problems. In this case, students were asked to use this model to make predictions about how the introduction of lionfish (considered an invasive species in some places) might alter the ecosystem.
Vole-Mustelid model
Viktoriia Radchuk Harry Peter Andreassen Rolf Anker Ims | Published Monday, October 07, 2019The model reflects the predator-prey mustelid-vole population dynamics, typically observed in boreal systems. The goal of the model is to assess which intrinsic and extrinsic factors (or factor combinations) are needed for the generation of the cyclic pattern typically observed in natural vole populations. This goal is achieved by contrasting the alternative model versions by “switching off” some of the submodels in order to reflect the four combinations of the factors hypothesized to be driving vole cycles.
An Individual-Based Mechanistic Model of Mussel Bed Boundary Formation and Intensity
Matthew Schumm | Published Saturday, September 28, 2019Individually parameterized mussels (Mytilus californianus) recruit, grow, move and die in a 3D environment while facing predation (in the form of seastar agents), heat and desiccation with increased tide height, and storms. Parameterized with data collected by Wootton, Paine, Kandur, Donahue, Robles and others. See my 2019 CoMSES video presentation to learn more.
Hybrid fish-plankton model
Gudrun Wallentin Christian Neuwirth | Published Friday, October 28, 2016 | Last modified Sunday, January 29, 2017A hybrid predator-prey model of fish and plankton that switches dynamically between ABM and SD representations. It contains 6 related structural designs of the same model.
A multithreaded PPHPC replication in Java
Nuno Fachada | Published Saturday, October 31, 2015 | Last modified Tuesday, January 19, 2016A multithreaded replication of the PPHPC model in Java for testing different ABM parallelization strategies.
Peer reviewed PPHPC - Predator-Prey for High-Performance Computing
Nuno Fachada | Published Saturday, August 08, 2015 | Last modified Wednesday, November 25, 2015PPHPC is a conceptual model for studying and evaluating implementation strategies for spatial agent-based models (SABMs). It is a realization of a predator-prey dynamic system, and captures important SABMs characteristics.
Peer reviewed Ache hunting
Kim Hill Marco Janssen | Published Tuesday, August 13, 2013 | Last modified Friday, December 21, 2018Agent-based model of hunting behavior of Ache hunter-gatherers from Paraguay. We evaluate the effect of group size and cooperative hunting
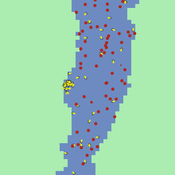
Landscape connectivity and predator–prey population dynamics
Jacopo A. Baggio | Published Thursday, November 10, 2011 | Last modified Saturday, April 27, 2013A simple model to assess the effect of connectivity on interacting species (i.e. predator-prey type)
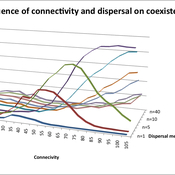
Varying effects of connectivity and dispersal on interacting species dynamics
Kehinde Salau | Published Monday, August 29, 2011 | Last modified Saturday, April 27, 2013An agent-based model of species interaction on fragmented landscape is developed to address the question, how do population levels of predators and prey react with respect to changes in the patch connectivity as well as changes in the sharpness of threshold dispersal?