About the CoMSES Model Library more info
Our mission is to help computational modelers develop, document, and share their computational models in accordance with community standards and good open science and software engineering practices. Model authors can publish their model source code in the Computational Model Library with narrative documentation as well as metadata that supports open science and emerging norms that facilitate software citation, computational reproducibility / frictionless reuse, and interoperability. Model authors can also request private peer review of their computational models. Models that pass peer review receive a DOI once published.
All users of models published in the library must cite model authors when they use and benefit from their code.
Please check out our model publishing tutorial and feel free to contact us if you have any questions or concerns about publishing your model(s) in the Computational Model Library.
We also maintain a curated database of over 7500 publications of agent-based and individual based models with detailed metadata on availability of code and bibliometric information on the landscape of ABM/IBM publications that we welcome you to explore.
Displaying 7 of 7 results coexistence clear search
Peer reviewed The Andean Resource Management Model (ARMM)
Olga Palacios | Published Tuesday, January 20, 2026ARMM is a theoretical agent-based model that formalizes Murra’s Theory of Verticality (Murra, 1972) to explore how multi-zonal resource management systems emerge in mountain landscapes. The model identifies the social, political, and economic mechanisms that enable vertical complementarity across ecological gradients.
Built in NetLogo, ARMM employs an abstract 111×111 grid divided into four Andean ecological zones (Altiplano, Highland, Lowland, Coast), each containing up to 18 resource types distributed according to ecological suitability. To test general theoretical principles rather than replicate specific geography, resource locations are randomized at each model initialization.
Settlement agents pursue one of two economic strategies: diversification (seeking resource variety, maximum 2 units per type) or accumulation (maximising total quantity, maximum 30 units). Agents move between adjacent zones through hierarchical decision-making, first attempting peaceful interactions—coexistence (governed by tolerance) and trading (governed by cooperation)—before resorting to conflict (theft or takeover, governed by belligerence).
The model demonstrates that vertical complementarity can emerge through fundamentally different mechanisms: either through autonomous mobility under political decentralization or through state-coordinated redistribution under centralization. Sensitivity analysis reveals that belligerence and economic strategy explain approximately 25% of outcome variance, confirming that structural inequalities between zones result from political-economic organization rather than environmental constraints alone.
As a preliminary theoretical model, ARMM intentionally maintains simplicity to isolate core mechanisms and generate testable hypotheses. This foundational framework will guide future empirically-calibrated versions that incorporate specific archaeological settlement data and geographic features from the Carangas region (Bolivia-Chile border), enabling direct comparison between theoretical predictions and observed historical patterns.
Individual-based model to "Intraspecific trait variation in personality‐related movement behavior promotes coexistence" (Milles et al., 2020)
Alexander Milles | Published Monday, June 22, 2020The community consequences of intra-specific trait variation (ITV) are a current topic in ecological research. The effects of ITV on species coexistence have, yet, not sufficiently been understood. With this individual-based model we analyzed the effect of intra-specific variation in movement by mimicking variation found in ground-dwelling rodents and analyzing how such variation affects inter-specific differences in competitive ability (i.e. foraging efficiency) and temporary coexistence. The movement algorithm and behavioral plasticity was adapted from existing algorithms and current ecological literature. As a measure for temporary coexistence, we analyzed the time until one of the species went extinct.
Managing networked landscapes: conservation in fragmented, regionally connected world
Jacopo A. Baggio Michael Schoon Sechindra Vallury | Published Monday, December 09, 2019Exploring how learning and social-ecological networks influence management choice set and their ability to increase the likelihood of species coexistence (i.e. biodiversity) on a fragmented landscape controlled by different managers.
Classical Swine Fever in wild boars
Volker Grimm Stephanie Kramer-Schadt Cédric Scherer Martin Lange Hans-Hermann Thulke | Published Friday, September 06, 2019The model is a combination of a spatially explicit, stochastic, agent-based model for wild boars (Sus scrofa L.) and an epidemiological model for the Classical Swine Fever (CSF) virus infecting the wild boars.
The original model (Kramer-Schadt et al. 2009) was used to assess intrinsic (system immanent host-pathogen interaction and host life-history) and extrinsic (spatial extent and density) factors contributing to the long-term persistence of the disease and has further been used to assess the effects of intrinsic dynamics (Lange et al. 2012a) and indirect transmission (Lange et al. 2016) on the disease course. In an applied context, the model was used to test the efficiency of spatiotemporal vaccination regimes (Lange et al. 2012b) as well as the risk of disease spread in the country of Denmark (Alban et al. 2005).
References: See ODD model description.
Kulayinjana
Christophe Le Page Arthur Perrotton Michel De Garine-Wichatitsky Barry Bitu Killion Koyisi Ferdinand Mwamba Cephus Ncube Victor Ncube Siphusisiwe Ndlovu Raphael Ngwenya Ambu Nyathi Fumbane Nyathi Patrick Sibanda Zenzo Sibanda | Published Monday, October 03, 2016a computer-based role-playing game simulating the interactions between farming activities, livestock herding and wildlife in a virtual landscape reproducing local socioecological dynamics at the periphery of Hwange National Park (Zimbabwe).
Managing Connectivity: insights from modelling species interactions accross multiple scales in an idealized landscape
Marco Janssen Michael Schoon Jacopo A. Baggio Kehinde Salau | Published Monday, October 01, 2012 | Last modified Monday, January 20, 2014The model objective’s is to explore the management choice set to uncover which subsets of strategies are most effective at maximizing species coexistence on a fragmented landscape.
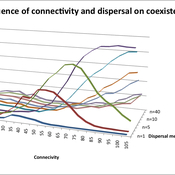
Varying effects of connectivity and dispersal on interacting species dynamics
Kehinde Salau | Published Monday, August 29, 2011 | Last modified Saturday, April 27, 2013An agent-based model of species interaction on fragmented landscape is developed to address the question, how do population levels of predators and prey react with respect to changes in the patch connectivity as well as changes in the sharpness of threshold dispersal?